How can you overcome the limits of FRET based protease activity assays for MMPs, ADAMs, Thrombin, Furin and Factor Xa?
Usually screenings for inhibitors of diverse protease activities are conducted with FRET or FRET-like assays, in which the protease substrate has to be coupled to a fluorophore and a quencher.
These assays use relatively short peptide substrates and often only the core of the specific protease recognition site can be exposed to the protease of interest. This can result in a number of false positives and negatives because the recognition site might not be highly selective. Furthermore these FRET substrates often show limited stability.
Fluorophores commonly used are furthermore characterized by a low level of photo stability, and tend to have short wavelengths which can result in higher backgrounds due to interfering fluorescence. Thus the signal-to-noise ration is sometimes on the borderline.
Another challenge with FRET based and FRET-like assays can be a high level of pH sensitivity and a low solubility of the substrates in aqueous solution. So, what’s the solution?
The alternative – Non-FRET EnSens Techology
Enzium have recently released a new activity assay for proteases like MMP-2, 9, 14, and 25, ADAM10 and 17, Furin, Thrombin and Factor Xa.
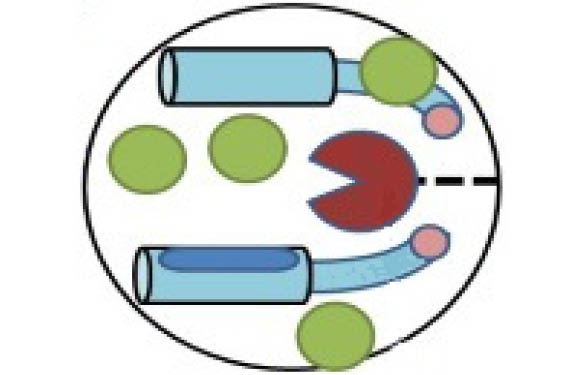
These assays are based on a non-FRET technology and make use of a protein substrate expressed in E. coli which is a modular dumbbell comprising two domains separated by a linker that contains a highly specific protease recognition sequence. One domain is the dye-binding domain, and the other is a blocking domain. When the linker is cleaved by a protease, separating the two domains, the binding domain is free to bind dye, constraining it and causing it to fluoresce (see Figure 1.).
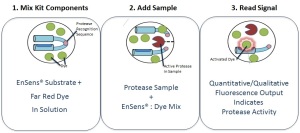
The benefits of this technology surpass all limitations of FRET and FRET-like assays:
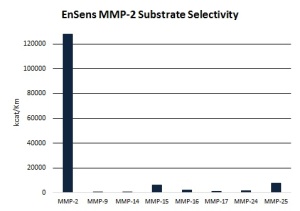
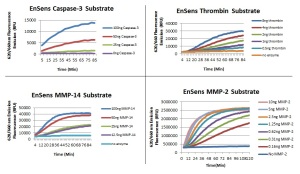
- The substrate can incorporate full recognition site sequences – thus the selectivity of the assays is superior (see Figure 2.)
- The substrate is modular, allowing easy swap of protease recognition sites – even customized substrates can be obtained if you know the recognition site of your protease of interest!
- The substrate is an scFv-based protein (single-chain variable fragment, an artificially generated antibody fragment) which confers high stability in vitro and in vivo.
- The EnSens se-Red fluorophore is resistant to photobleaching up to 48 hours
- The long emission wavelength (665nm) reduces interference from other assay components
- The assay works stably in wide range of pH, salt, & solvent conditions (see Figure 3.)
- Substrate and fluorophore are separate in the systems, so the fluorophore is constantly renewed in solution
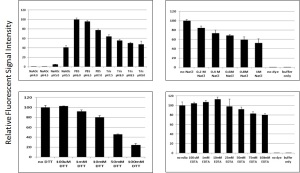
Figure 3: EnSens® is stable in varying conditions often used in biological sample and enzyme kinetic assays - The system is characterized by high signal-to-noise ratios: 6-20x
- The read-out can be done in standard plate or cuvette fluorimeters
- The EnSens assay gives repeatable and reproducible kinetic activity curves from reagent to reagent (see Figure 4.)
- The same assay principle can be applied to flow cytometry experiments, standard and 3D cell culture microscope (customized option).
Do you think this could help your research? Leave your questions or comments below!