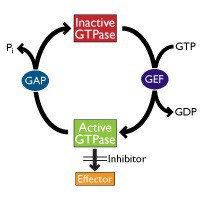
In this post, I invite you to discover some state-of-the art methods to measure the activation of the most prominent small G proteins. In future posts, we’ll then be looking at methods to measure the activity of Guanine nucleotide exchange factors (GEFs) which stimulate the release of GDP to allow the binding of GTP (which activates the small G protein) and of GTPase-activating proteins (GAPs) which stimulate the GTPase activity of small G proteins to inactive them (see Fig. 1).
The Ras superfamily of small G proteins consists of more than 150 members which can be divided into five main families – Ras, Rho, Ran, Rab and Arf. Small G proteins cycle between the inactive, GDP-bound form and the active, GTP-bound form (Fig. 1).
The Rho subfamily consists of proteins like RhoA, Rac1, and Cdd42. These proteins have been shown to be involved in the regulation of actin dynamics, thus playing a crucial role in processes like cell movement, intracellular transport, and organelle development. While RhoA affects actin stress fibers, Rac1 exhibits effects on lamellipodia and Cdc42 on filopodia.
The Ras subfamily consists of Ras, Ral, Rit, Rap, Rheb, and Rad. Most prominent is Ras as all three human Ras genes (H-Ras, K-Ras, and N-Ras) represent well-known oncogenes in cancer. In ap. 20-25% of all human tumors mutated forms of Ras can be found. Thus Ras is seen as an important target for anti cancer drug development.

Approaches for measuring Small G Protein activation
The classical way to measure the activation of small G proteins (i.e. the transformation of the GDP-bound form to the GTP-bound form) is a pull-down assays. The assays make use of effector proteins which specifically bind to the activated forms of small G proteins. Coupled to beads these proteins can be used to pull-down the activated small G protein from a cell lysate. Lysates from control cells can be compared to lysates from treated or modified cells.
An even more convenient assay format based on the same principle is the G-LISA technology developed by Cytoskeleton Inc. In this technology, the effector proteins which selectively bind to activated small G proteins are coupled to 96 well plates, making it possible to run an ELISA-like activation assay (for typical G-LISA results see Fig. 2).
To get a quick overview about both techniques, you might like to watch the video below.
Which kind of assays are available for which targets?
Pull-down assays are available for:
Arf1 – Arf6 – Cdc42 – Rac1 –RalA – Ras – RhoA
G-LISA assays are available for:
Arf1 – Arf6 – Cdc42 – Rac1 – Rac1/2/3 – RalA – Ras – RhoA
How to start in the most cost-effective way?
Note that some of these kits are available as trial sizes for cost effective first tests.
If you’re not sure about which of the major small G proteins (RhoA, Rac1, or Cdc42) are affected in your experiments, an interesting option you might consider starting with is the Combo kit, containing pull-down assays for RhoA, Rac1, as well as Cdc42.
Any question or comments? Feel free to leave them below!