Mitochondria keep cells “healthy” in their environment. In this post, we’ll take a look at a new fluorescent assay for the monitoring of mitophagy, a process aimed at clearing damaged or superfluous mitochondria by autophagy, in mammalian cells.
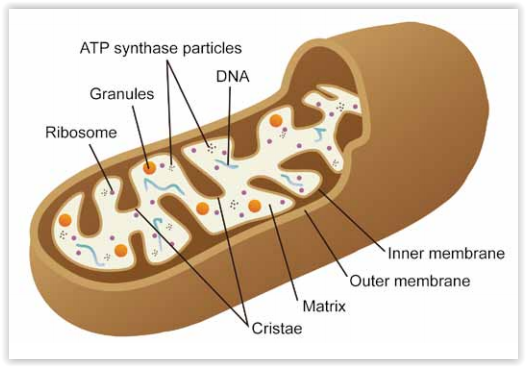
Mitochondria are cellular organelles with crucial functions in cell homeostasis (energy conversion, apoptosis, cell differentiation …). As for the cytoskeleton (see “Actin and Tubulin dynamics research studies“), mitochondria are subject to dynamic processes that maintain the pool of mitochondria functional . These events ((biogenesis and fusion & fission) ensure an optimal exchange of organelle contents in the cells, together with the recycling of “damaged” mitochondria (displaying a depolarized mitochondrial membrane potential for example) into functional mitochondria.

Damaged mitochondria are mainly recycled back and degraded by mitophagy. Mitophagy selectively eliminates dysfunctional mitochondria (often linked with Reactive Oxygen Species and DNA damage) by sequestering them into autophagosomes that are then fused to lysosomes for further degradation. The selective autophagic recognition events are notably mediated by the Pink-1-Parkin cell signaling pathway. Mitophagy has been described to be involved in aging and neurodegenerative disorders (e.g. Parkinson’s and Alzheimers’s diseases) but also in drug-induced tissue injuries and inflammation.
Experimental methods allowing the analysis of mitophagy are quite diverse (e.g. microscopy techniques, immuno-assays specific for mitochondrial markers…). Recently, Dojindo has developed a fluorescent assay kit to accurately monitor mitophagy in mammalian cells: the Mitophagy detection kit.
Dojindo’s new Mitophagy detection kit
The Mitophagy detection kit (cat. nr D535-08) is composed of Mtphagy Dye (for detecting mitophagy) and Lyso Dye (for detecting lysosome).
During the the assay, the Mtphagy Dye accumulates in mitochondria with a weak fluorescence. When Mitophagy is induced, the damaged mitochondria fuses to lysosome leading to a high fluorescent signal emitted by the Mtphagy Dye. The fusion of Mtphagy Dye–labeled mitochondria with lysosome is then tracked by the Lyso Dye which confirming “mitochondria-lysosome” colocalization. (see experimental data below on HeLa cells).

Fluorescence microscopy allowing “mitochondria-autophagosome” or “mitochondria-lysosome” colocalization is not something new. Nevertheless, this is the first time that an assay kit integrates 2 fluorescent dyes that have been optimized for mitophagy detection.
If you are looking for other mitochondrial fluorescent dyes, you might consider the following ones from Genecopoeia / Applied BioProbes:
- MitoView Red (a.k.a. as MitoTracker® Red CMXRos*) is an red-fluorescent dye (Ex/Em (nm): 580/600) that stains mitochondria in live cells. Its accumulation is dependent upon membrane potential. The dye is well-retained after aldehyde fixation.
- MitoView Orange (a.k.a. as MitoTracker® Orange CMTMRos*) is an orange-fluorescent dye (Ex/Em (nm): 554/575) that stains mitochondria in live cells and its accumulation is also dependent on the membrane-potential. The dye is well-retained after aldehyde fixation.
- JC-1 stains mitochondria in living cells in a membrane potential-dependent fashion (Ex/Em (nm): 514/529; 585/590). JC-1 exists as a monomer at low concentrations and yields green fluorescence, similar to fluorescein. At higher concentrations, the dye forms J-aggregates that exhibit a broad excitation spectrum and an emission maximum at ~590 nm. These characteristics make JC-1 a sensitive marker for mitochondrial membrane potential. JC-1 is particularly useful for apoptosis studies. In apoptotic cells, the dye stays in the cytoplasm and fluoresces green.
Interested in using other mitochondrial research tools like this?

Take a look at our selection here, or leave your comments or questions below:
- New SiR-fluorogenic probes for monitoring lysosome by multicolour Live Cell Imaging
- ROS related assay kits
- Customizable E3-ubiquitin Ligase assay kits and small molecules known to be inhibitors of proteosomal & Ub-related enzymes
- Antibodies raised against proteins involved in mitophagy and mitochondrial research
- Anti-Beclin 1 and anti-AMBRA1 polyclonal antibodies validated for WB analysis (Bioworld Technology)
- Anti-Parkin polyclonal for WB (1:500~1:1000), IHC (1:50~1:100), ELISA (1:10,000) (Assay Biotech)
- Anti p-Parkin (Phospho-Ser131) for phosphorylation studies by IHC or ELISA (Assay Biotech)
- Anti-PINK1 Mouse monoclonal (Clone 38CT20.8.5) for WB (1:100~1:500), IHC (1:50~1:100), IF (1:25) (Abnova)
- 10 Mitochondrial marker antibodies
Sources:
- Ding WX, Yin XM “Mitophagy: mechanisms, pathophysiological roles, and analysis” (2012) Biol Chem. Jul;393(7):547-64. doi: 10.1515/hsz-2012-0119.
- Gomes LC, Scorrano L “Mitochondrial morphology in mitophagy and macroautophagy” (2013) Biochim Biophys Acta. 1833(1):205-12. doi: 10.1016/j.bbamcr.2012.02.012.
- Durcan TM, Fon EA “The three ‘P’s of mitophagy: PARKIN, PINK1, and post-translational modifications” (2015) Genes Dev. May 15; 29(10): 989–999. DOI: 10.1101/gad.262758.115.
- Zhang CW et al. “Parkin Regulation and Neurodegenerative Disorders” (2016) Front Aging Neurosci. Jan 12;7:248. DOI: 10.3389/fnagi.2015.00248.
* Trade Mark of Molecular Probes