In the summer of 2013, my colleague Frederic Dubor, who is in charge of our services lab, mentioned to me that he had just been asked by 3 clients within a very short time-frame, whether our laboratory would be able to run histone binding studies for them. In fact, it wasn’t about binding to single full length histones, but all 3 clients had asked for multiplex arrays, meaning they were interested in measuring binding of either antibodies (epitope mapping) or epigenetic readers (bromodomains and methyl readers) to a set of histone derived peptides, without and with a number of modifications such as acetylation and methylation,
At that time, neither Frederic nor I could help – but these requests did make us search for a supplier offering arrays which would meet these needs. Half a year later, we were pleased to announce a new cooperation with Epicypher, one of the market leaders in epigenetic proteomics.
But I’m running ahead a little quickly… I’d first like to summarize the potential applications of the arrays we were not able to offer one year ago.
So, what precisely were researchers looking for when asking to “run histone binding studies”?
1. The determination of the exact modification on a specific histone recognized by an antibody raised against histones. In fact, this could be meant as a strigent quality control experiment for an antibody to be used to recognize specific histone modifications. Furthermore, such a specific antibody could be used to block out the binding of any other protein to the histone target sequence. As the 4 histones which make the nucleosome’s histone octamer (H2A, H2B, H3, and H4) can be epigenetically modified by a number of chemical reactions, such as acetylation, methylation, phosphorylation, ubiquitination, sumoylation, and ADP Ribosylation, theoretically thousands of modification patterns can be found on histones.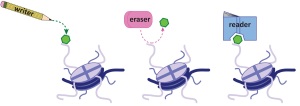
2. The determination of specific binding sites of epigenetic “readers”, such as bromodomains (recognizing acetylated sites) and methyl readers. These “readers” can be domains of “writers” (enzymes modifying histones, e.g. methyl transferases and acetylases) or “erasers” (e.g. demethylases and deacetylases). Thus, these modifying enzymes can be targeted to their sites of action on histones.
3. Furthermore, a third application field for peptide arrays containing diverse modified and non modified histone sequences could be the analysis of the substrate specificity of entire histone modifying enzymes.
All these applications could help in answering questions in research fields and diseases which are linked to epigenetic histone modifications such as cancer, aging, asthma, diabetes, cardiovascular diseases, neurodegeneration, autoimmune disorders, and developmental disorders.
We found arrays to answer all these questions: Precise histone binding studies are now possible with EpiCypher’s EpiTitan Histone Peptide Arrays
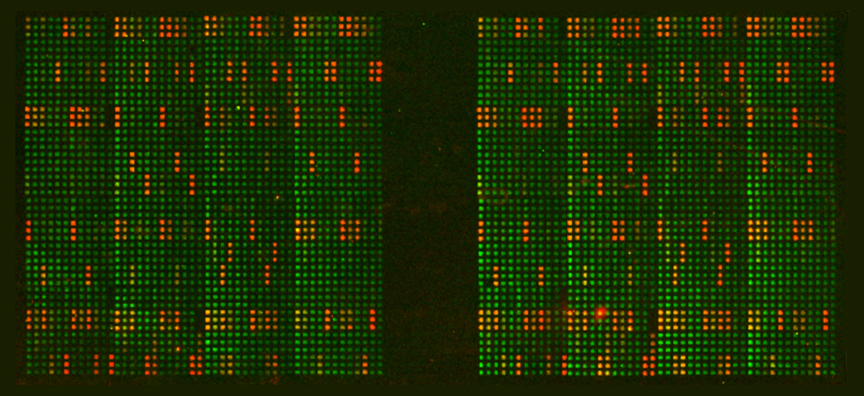
The EpiTitan™ Histone Peptide Arrays represent the next generation in histone peptide arrays, and are the gold standard against which all other histone peptide arrays are judged.
The arrays feature hundreds of highly purified and well-characterized histone peptides spotted onto the substrate. EpiTitan™ Histone Peptide Arrays are a dramatic improvement over arrays in which the peptides are synthesized on the substrate (SPOT synthesis), a technique that leads to peptides on the array that are of unknown quantity and quality. All the peptides on the EpiTitan™ Histone Peptide Arrays are purified by HPLC and validated by mass spectrometry prior to spotting.
The technique used in this array is quite easy.
- Biotinylated peptides are spotted 12 times each per array on a surface covered with streptavidin to catch the peptides.
- Each array slide comes with two subarrays, thus two samples per slide can be evaluated.
- The protein to be investigated is added and binds to its specific target sequence and modification pattern.
- The detection of binding either starts with an antibody against the protein of interest or an anti tag antibody, should the protein carry the respective tag, and can be visualized using fluorescence scanning or standard ECL techniques – by the way the EpiTitan™ Histone Peptide Array is the only array available on the market which supports both detection methods.
- Protocols for qualitative as well as quantitative analysis of the data are provided.
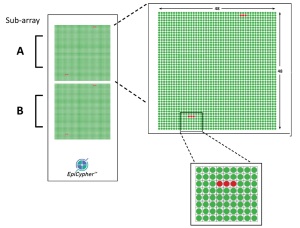

Design of the Arrays and experimental steps
Interested in more information or using EpiTitan™ Histone Peptide Arrays yourself?
Just get in touch if you’d like to check the list of histone peptides and the user manual. I’ll be happy to get back to you to tell you more about these Histone arrays and companion reagents (Epigenetic readers and erases or bioactive small molecules…).
[contact-form to=’Ali.el.baya@tebu-bio.com’ subject=’Please contact me for further information about the Epititan histone peptide array’][contact-field label=’Name’ type=’name’ required=’1’/][contact-field label=’Email’ type=’email’ required=’1’/][contact-field label=’Comment’ type=’textarea’ required=’1’/][/contact-form]
While reading this blog you were maybe already thinking about how you could apply this array in your research project…
Then I have good news for you. Our partner Epicypher just started a grant program – please have a look at our previous blog: 5 fully funded Epigenetic Grants offered by Epicypher.
The principle of the grant program is very easy – you supply a brief outline of a study which involves the role of histone modifications in physiological or pathological processes – Epicypher supplies arrays free of charge to five applicants chosen by an independent scientific committee.